Guide to MetaCyc
Contents
2 MetaCyc Overview
2.1 Motivations
2.2 Database Contents
2.3 Query and Visualization
2.4 The MetaCyc Data Universe
2.5 Linking to MetaCyc
2.6 Development
2.7 Underlying Software
4 MetaCyc Curation
4.1 Information Types Captured During the MetaCyc Curation Process
4.1.1 Pathways
4.1.2 Reactions
4.1.3 Enzymes and Enzyme Complexes
4.1.4 Genes
4.1.5 Compounds
5 Taxonomic Designations for Pathways
6 Release Process, Frequency, and History
6.1 MetaCyc Release Procedures
10 Comparison of MetaCyc to other Pathway Databases
10.1 KEGG
10.2 EAWAG Biocatalysis/Biodegradation Database
10.3 Reactome
11 The MetaCyc Team
11.1 Current Contributors
11.2 Past Contributors
12 Submitting Pathways for Incorporation into MetaCyc
12.1 How to Ensure that You and Your Organization Receive the Appropriate Credit
12.2 How to Send Pathways to MetaCyc
13 MetaCyc Publications
13.1 Additional Publications
1 Introduction
This guide provides additional information on the MetaCyc database (DB) beyond that found in other MetaCyc publications [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11], to help users of the database understand its contents in more depth.
MetaCyc is a member of the BioCyc collection of Pathway/Genome Databases. In contrast to all other members of that collection, which are organism-specific DBs, MetaCyc is a multiorganism DB. The other BioCyc databases describe the metabolic network and genome of a single organism, and mix experimentally determined pathways with computationally predicted pathways. MetaCyc contains experimentally elucidated pathways only; one goal of the MetaCyc project is for MetaCyc to contain a representative example of as many experimentally-determined metabolic pathways as possible.
MetaCyc does not seek to model the complete metabolism of any particular organism, which is the role of individual BioCyc DBs. Instead, MetaCyc serves as a general reference on metabolic pathways and enzymes. MetaCyc is also used as a high-quality reference DB for predicting metabolic pathways in other organisms. Scientists use MetaCyc for a broad range of tasks, such as finding enzymes for metabolic engineering projects, learning about the possible metabolic fates of specific compounds, identifying metabolites based on mass spectrometry data, or finding a comprehensive description for complex pathways.
For questions that require information about the complete genome, proteome, or metabolic network of a particular organism, users are advised to consult the appropriate organism-specific PGDB. For example, MetaCyc contains 21 pathways and 112 enzymes that have been experimentally studied in Staphylococcus aureus. In contrast, the BioCyc Staphylococcus aureus database contains 201 pathways (most of which are computationally predicted), plus the entire genome and proteome of that strain.
2 MetaCyc Overview
MetaCyc is a database of non-redundant, experimentally elucidated metabolic pathways and enzymes. It also contains reactions, chemical compounds, and genes. It stores predominantly qualitative information rather than quantitative data, although it does contain some quantitative data such as enzyme kinetics data. “MetaCyc” is pronounced “met-a-sike”. It sounds like “encyclopedia”.
A unique property of MetaCyc is that it is curated[def] from the scientific experimental literature according to an extensive process [4], such that:
3,443 different organisms are represented, with the majority of pathways occurring in microorganisms and plants
3,128 metabolic pathways are stored, with 18,819 enzymatic reactions and 76,283 associated literature citations
MetaCyc stores the full complement of reactions that have been assigned EC numbers by the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB)
MetaCyc stores thousands of additional enzyme-catalyzed reactions that have not yet been assigned an EC number
2.1 Motivations
The MetaCyc mission is to serve a broad community of researchers from genetics, molecular biology, microbiology, biochemistry, genomics, metabolomics, bioinformatics, metabolic engineering, and systems biology in support of the following tasks:
- Support computational metabolic reconstruction and modeling
-
One of MetaCyc’s primary applications is to serve as a reference
database for computationally predicting the metabolic network
of an organism from its annotated genome, such as by the PathoLogic
component of Pathway
Tools [example] .
Those qualitative metabolic reconstructions can be converted to metabolic models using the
MetaFlux component of Pathway Tools.
- Provide an encyclopedic reference on pathways and enzymes
-
MetaCyc is used as a readily accessible source of up-to-date,
literature-curated information on metabolic pathways and enzymes
for basic research and genome analysis,
as well as for educational purposes. [example]
- Support metabolic engineering
-
Metabolic engineers use MetaCyc as an encyclopedia of metabolic
pathways and enzymes that may be genetically engineered into
an organism to alter its metabolism. [example]
- Support metabolomics studies
-
MetaCyc’s rich content of metabolites with full structure and
monoisotopic mass data
makes it a useful resource for the identification of metabolites from mass spectrometry data.
2.2 Database Contents
MetaCyc stores pathways involved in both [Primary metabolism] and [Secondary metabolism].
MetaCyc’s metabolites, enzymes, enzyme complexes, and genes are not limited to those associated with these pathways; the database contains many enzymes and reactions not associated with pathways.
MetaCyc is extensively linked to other biological databases [8] including protein and nucleic-acid sequence databases and chemical compound databases.
While MetaCyc itself does not contain protein or gene sequence data, protein sequences are easily retrievable for MetaCyc enzymes that have a link to UniProt. To retrieve this information, Click on the "Show Sequence at UniProt" command in the Operations menu.
2.3 Query and Visualization
MetaCyc data can be browsed and queried in multiple ways. For pathways, proteins, reactions and compounds, the MetaCyc site supports:
Text-based searching: simple text searches are possible by typing in the quick search box at the top of the page. You can hover the mouse inside the box to see a detailed description of the available options [example]
Direct queries: extensive search capabilities are provided via a user-friendly search interface accessible via the Tools → Search commands [example]
Advanced search: an advanced search option that lets users build a complex query is available via the Tools → Advanced Search command
Browsing using ontologies: this option is useful when one wants to search by proceeding from a general category to a specific instance. It is accessible via the commands Tools → Browse Pathway Ontology and Tools → Browse Compound Ontology and Tools → Browse Enzyme Commission Ontology [example]
Extending information to other databases: every object in MetaCyc enables quick searching for it in a specific organism or a set of organisms via the Operations menu (right side) commands Show This Object in Another Database and Search for this Object in Multiple Databases. Comparison features combine MetaCyc with other BioCyc databases to provide additional ways for viewing data. Examples for Cross-Species comparisons include:
Comparing specific pathways between two or more organisms [example]
Comparing the genomic maps of two or more organisms [example]
2.4 The MetaCyc Data Universe
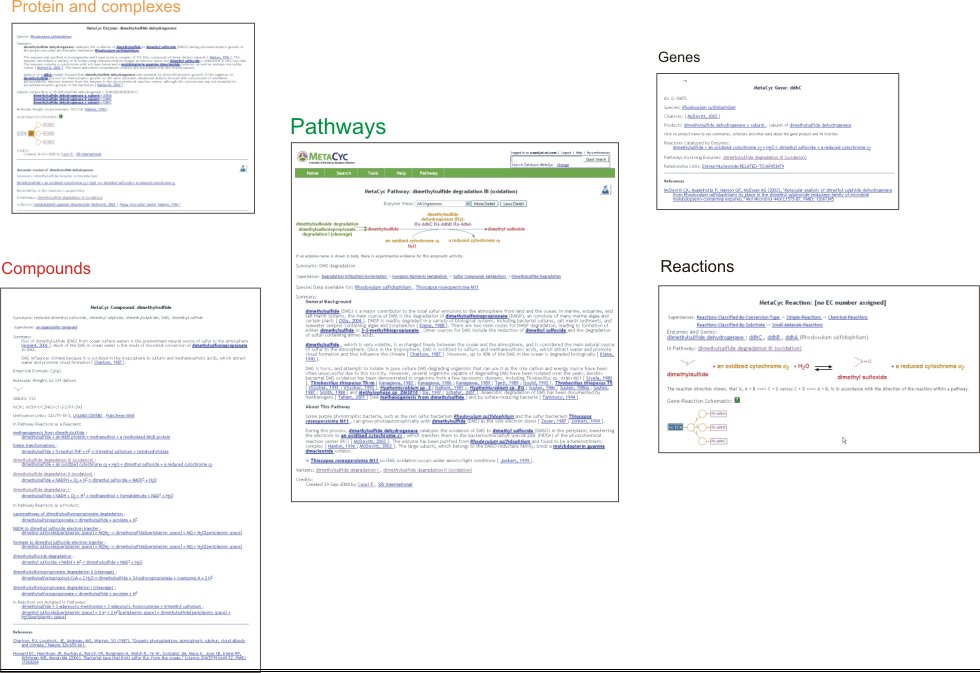
MetaCyc inter-relates information about pathways, reactions, compounds, proteins, and genes. Each object name is typically a hyperlink to the page describing that object. For example, while looking at a pathway page, clicking on a compound, reaction arrow, protein name, or gene name will navigate to those object’s pages, making it extremely easy for the user to navigate among the different database’s objects.
2.5 Linking to MetaCyc
Users are encouraged to link their Web site or application to MetaCyc as described here.
2.6 Development
Since its beginning in 1998, MetaCyc’s data have been gathered from a variety of literature and on-line sources. MetaCyc is currently curated by a single full-time curator at SRI International. Some of the data in MetaCyc have been curated via collaborative projects between SRI International and the Marine Biological Laboratory, the Carnegie Institution’s Department of Plant Biology, and the Boyce Thomson Institute for Plant Research.
2.7 Underlying Software
Curation and navigation of MetaCyc is performed using the Pathway Tools platform — which can be obtained here.
3 MetaCyc Availability
MetaCyc is available via a BioCyc subscription in several different forms to facilitate different uses of the data:
The MetaCyc data are available through the MetaCyc Web site for interactive search and visualization.
The MetaCyc data can be downloaded as a set of data files. These files can be parsed and queried using languages like Perl, or they can be loaded into other database systems. Click here for more information about the flat files, or you can download them from here.
The Pathway Tools software includes MetaCyc and can be downloaded and installed on computers at your site, with the following advantages:
It provides some functionality that is not provided by the MetaCyc Web site
It usually runs more quickly than the Web site and does not depend on an internet connection
It supports programmatic querying of MetaCyc using APIs in the Python, Java, Perl, and Lisp languages
It supports running an equivalent of the BioCyc Web site on your intranet
4 MetaCyc Curation
Curation is the process of manually refining and updating a bioinformatics database. The MetaCyc project uses a literature-based curation approach in which database contents are extracted in a step-wise manner from evidence in the experimental literature, as depicted below.
The curation procedures that MetaCyc curators follow are described in the Curator’s Guide to Pathway/Genome Databases.
MetaCyc data are derived from primary literature, review articles, patent applications, and from external databases.
For certain organisms, some of the data within MetaCyc have been directly imported from other databases which we consider to be the authoritative sources of data on those organisms:
 |
4.1 Information Types Captured During the MetaCyc Curation Process
Note that not all objects contain all of the information types listed here; rather, this list describes the potential types of information for each object type.
4.1.1 Pathways
Pathways include a mini-review summary that usually includes:
Background information about the metabolite(s) being synthesized or degraded
Relationship to other pathways
Background information about the research leading to establishing the pathway
Whether the pathway occurs naturally or is engineered
The expected taxonomic distribution of the pathway
Experimental evidence for the pathway
Contradictory evidence
Citations to the literature from which the information was derived
Other collected data include:
Synonyms
Taxonomic range
Key reactions
4.1.2 Reactions
EC number(s)
Free energy (mostly calculated from energy of formation of participating metabolites)
Whether this is a hypothetical reaction
Whether the reaction occurs spontaneously
Whether the reaction is/can be balanced (some reactions, such as reactions describing extension of polymers, cannot be balanced)
4.1.3 Enzymes and Enzyme Complexes
Most enzymes include a mini-review summary that covers:
Background information about the research conducted about the enzyme
Whether experimental evidence is available, and of what type
Whether the enzyme exists as a monomer or a complex
Experimental evidence
Isoforms
Substrate specificity
Tissue type and subcellular location
Citations to the literature from which the information was derived
Other collected data:
Common name of the enzyme and synonym(s)
Subcellular location
Molecular weight (KDa)
pI
Name(s) of cofactor(s), activator(s) and inhibitor(s)
Alternative substrates
Km, Vmax, Kcat, specific activity
Optimum pH
Optimum temperature
Directionality of the catalyzed activity
A link to an external database that contains sequence information, such as UniProt or the NCBI Protein database
4.1.4 Genes
Common name of the gene and synonym(s)
Accession numbers
Citations
A link to an external database that contains sequence information, such as Entrez Nucleotide or Gene.
4.1.5 Compounds
Recommended name and synonyms of the compound
Compound structure
Ontology (when available)
Free energy of formation
Link(s) to external databases such as ChEBI, KEGG, ChemSpider, PubChem
Compound structures are obtained either from the primary literature or from public compound structure databases such as ChEBI and ChemSpider. The structures are edited using the Marvin software to provide a consistent look and to reflect the most prevalent protonation state at pH 7.3. For more information about protonation, see Reaction Balancing and Protonation State in BioCyc at the Guide to The BioCyc Database Collection.
We would like to express our gratitude to Chemaxon for granting us a free license to their Marvin software.
5 Taxonomic Designations for Pathways
MetaCyc pathways are labeled with the name of one or more taxa in which wet-lab experiments have indicated that the pathway is present. These taxonomic designations are present on the pathway page in a line labeled “Some taxa known to possess this pathway include,” and include species names, species and strain names, and occasionally names of higher taxa such as genus names, e.g., Pseudomonas. When a high-level taxon, such as a genus, is present as a pathway label, the interpretation is that experimental evidence suggests that the pathway is present in all members of that taxon.
The “number of organisms” row in the MetaCyc statistics indicates the total number of different organisms that are referenced in the database. These could be listed in the taxonomic designations of pathways, but could also be references by enzymes that do not participate in pathways, or simply be mentioned in pathway/enzyme summaries. There is wide variation in how many pathways a given taxon contributes to MetaCyc, with some taxa contributing only a single pathway, and other taxa contributing more than 100 pathways. The taxonomic distribution of MetaCyc pathways is summarized here: [Pathway Taxonomic Distribution] .
To query MetaCyc pathways by species:
Web version: Select MetaCyc as the current database, then Tools → Search → Search Pathways then expand Search/Filter by organism
Desktop version: Select MetaCyc as the current database, then Pathway → Search by Organism
MetaCyc pathway pages also specify an “Expected taxonomic range,” which lists the taxonomic groups in which this pathway is expected to occur. This information is useful for pathway prediction.
6 Release Process, Frequency, and History
New versions of MetaCyc are released 3–4 times per year.
One release is a “minor” release, that is, data releases are made available on the MetaCyc.org Web site and in downloadable data files.
Two releases are “major” releases, which include the updates available in minor releases, plus new versions of Pathway Tools.
A detailed history of the enhancements to MetaCyc in each MetaCyc release is available here. This page also contains statistics on the changes in the content of MetaCyc over time.
6.1 MetaCyc Release Procedures
The MetaCyc staff perform the following operations as part of each MetaCyc major release (some are performed for minor releases as well):
Import data from the latest version of the ExploreEnz DB into MetaCyc to ensure MetaCyc contains the latest EC number information.
Import the latest version of the NCBI Taxonomy DB [12] into Pathway Tools.
Import the latest version of the GO database into Pathway Tools.
Update MetaCyc links to external DBs.
Compute InChI strings for MetaCyc metabolites using the InChI software [13].
Compute protonation states for MetaCyc metabolites using ChemAxon’s Major Microspecies plugin.
Compute Gibbs free energies for MetaCyc metabolite using an SRI software tool.
Compute reaction atom mappings using an SRI software tool [14].
Run many quality assurance programs including the Pathway Tools Consistency Checker; investigate and repair data glitches uncovered by these tools. Examples:
Check that reactions are balanced
Check for duplicate chemical compounds
Check chemical structures for invalid bonds and elements
Validate all GO terms as defined within the latest version of GO
Validate all internal database relationships as referring to valid database objects (e.g., subpathways listed for a given pathway must be valid database objects)
Ensure that slot values meet range and cardinality constraints (e.g., values of the pI slot must be between 0–14).
Validate literature references, e.g., check that PubMed IDs refer to actual PubMed entries
Check syntax of external database links
Check syntax of HTML embedded in comments
Update DB and software documentation and web pages.
7 BLASTing Against MetaCyc
A common early step in performing pathway analysis of genomes and metagenomes is to associate protein sequences to MetaCyc reactions. The Pathway Tools software infers such associations by using EC numbers, enzyme names, and Gene Ontology terms within protein annotations. Such annotations might be inferred using a variety of sequence-analysis methods.
To aid researchers in associating sequences to MetaCyc reactions, MetaCyc enzymes
that have a link to UniProt contain protein sequence information. It is possible
to perform BLAST searches against MetaCyc proteins with sequence information
using the "BLAST Search" command under the Search menu. In addition, each release
of MetaCyc includes a file that associates MetaCyc reaction IDs with the
UniProt identifiers of enzymes known to catalyze those reactions. Note that
not all MetaCyc reactions have EC numbers (because many enzymes have not yet
been assigned EC numbers), therefore EC numbers are not a comprehensive mechanism
for associating sequences to reactions. The file is called uniprot‑seq‑ids.dat
and is included in the MetaCyc data file distribution.
8 Database Links
MetaCyc contains links to many other bioinformatics DBs. Some MetaCyc links are “unification links”, meaning they are links from an object in MetaCyc to an object in another DB that represents the same biological object. Other links are “relationship links”, meaning that they are links from an object in MetaCyc to an object in another DB that represents a related object, such as a link from a MetaCyc reaction to a PIR protein that catalyzes that reaction. Note that not all objects contain links to all of the databases listed here; rather, this list describes the potential links for each object type.
The following types of MetaCyc objects contain links to the following databases.
Pathways link to EcoCyc, the EAWAG Biocatalysis/Biodegradation Database, and the Soybase database.
Proteins contain unification links to UniProt, FlyBase, DictyBase, DIP, DisProt, STRING, EuPathDB, PhosphoSite, PRIDE, PROSITE, SMR, and MINT.
Proteins also contain relationship links to SMART, PANTHER, ProDom, PRINTS, CAZy, PDB, Pfam, and InterPro.
Reactions and EC numbers link to ENZYME, KEGG, BRENDA, Rhea, and ExplorEnz.
Chemical compounds link to ChEBI, PubChem, NCI Open Database, KEGG, Chemical Abstracts, Wikipedia, ChemSpider, DrugBank, BiGG and KEGG Glycan.
Genes link to NCBI Entrez nucleotide and gene databases, to TAIR (The Arabidopsis Information Resource) and to the Coli Genetic Stock Center.
All types of objects may contain literature citations, which link to PubMed when available or directly to the original publication via a DOI.
9 Data Sources
MetaCyc incorporates information that was obtained from the following sources:
- The primary literature
- Most of the data in MetaCyc is derived
from the primary literature. Every pathway and most enzymes are based on
articles published in peer-reviewed magazines. Many literature
citations are obtained via PubMed.
- The EC list
- The reactions in MetaCyc that have EC numbers were
originally defined by the Nomenclature Committee of the
International Union of Biochemistry and Molecular Biology
(NC-IUBMB), as seen in
the ExplorEnz database. Many
of the EC reactions were imported into MetaCyc via the
ENZYME database, an enzyme
nomenclature database based on the recommendations of the NC-IUBMB,
with the kind permission of the Swiss
Institute of Bioinformatics.
Subsequent to their import into MetaCyc, we have modified many of the EC reactions from the original IUBMB format. One main reason for the modifications is the fact that in MetaCyc the compounds that participate in the reactions are actually database objects, rather than strings of text. The ExplorEnz and ENZYME DBs suffer from some inconsistency so that the same compound may be referred to by different names in different reactions. In MetaCyc reactions the compound is represented by a reference to an underlying DB entry for the compound, and thus will always appear with the same common name. A second reason for modification is our attempt to provide a consistent format for the representation of reactions, in the form of “reactants ⇋ products”, which format is not always used by the IUBMB. For example, reaction EC 3.4.24.86, ADAM 17 endopeptidase, is defined by the IUBMB by the free text: “Narrow endopeptidase specificity. Cleaves Pro-Leu-Ala-Gln-Ala-|-Val-Arg-Ser-Ser-Ser in the membrane-bound, 26-kDa form of tumor necrosis factor alpha (TNF-alpha). Similarly cleaves other membrane-anchored, cell-surface proteins to ’shed’ the extracellular domains”. In MetaCyc the reaction is presented simply as “a TNF-α precursor + H2O → an active TNF-α + a peptide”, and the rest of the information is provided within the reaction comment. A third reason for the differences results from the fact that all MetaCyc compounds are protonated to a consistent pH (the cytosolic pH of 7.3), and all reactions are balanced for mass and charge by the addition of protons where applicable (most NC-IUBMB reactions do not consider protons).
- UniProt protein database
- Most of the polypeptides in MetaCyc are
linked to UniProt. UniProt data is
often used as the source for the enzyme’s molecular weight
(calculated from sequence), for links to DNA sequence databases, and
for synonyms of the enzyme and its gene. It is also used to help
determine the taxonomic distribution of enzymes.
- NCBI Taxonomy, Nucleotide and Gene databases
- The
NCBI taxonomy database is
integrated into MetaCyc, and all internal references to organisms
that are included in that taxonomy are based on the NCBI taxonomy
IDs. MetaCyc gene entries are most commonly linked to the NCBI
Nucleotide and Gene databases.
- ChEBI, KEGG Ligand and PubChem chemical structure databases
-
While most of the chemical structures in MetaCyc were obtained from
the original literature, many were obtained from other databases.
MetaCyc entries contain DB links to many other compound databases,
including ChEBI,
KEGG and
PubChem.
- The BRENDA enzyme database
- When predicting the taxonomic range
of pathways, MetaCyc curators occasionally use data from
BRENDA to estimate the
taxonomic range of individual enzymes.
10 Comparison of MetaCyc to other Pathway Databases
10.1 KEGG
A detailed comparison of KEGG and BioCyc was posted here in 2023: https://bioinformatics.ai.sri.com/biocyc/kegg-biocyc-comparison.pdf. Another comparison of KEGG and MetaCyc was published in 2013 [15].
KEGG contains two types of pathways: maps and modules.
KEGG maps are quite large and typically integrate reactions and pathways found in multiple species – they are therefore chimeric. KEGG maps are not found in their entirety in any one species. MetaCyc superpathways are similar to KEGG maps in that they comprise multiple sub-pathways, although unlike KEGG maps, most MetaCyc superpathways do occur in a single organism.
KEGG modules are smaller than maps, and are similar in size to individual MetaCyc pathways (although KEGG’s collection of modules is very incomplete because they are a relatively new development in KEGG).
MetaCyc pathways (and KEGG modules) are closer to true biological pathways than are KEGG maps because they attempt to model individual biological pathways from individual organisms. KEGG maps are typically 3–4 times larger than are KEGG modules and MetaCyc pathways because of the chimeric nature of KEGG maps. For example, KEGG map MAP00270 called “cysteine and methionine metabolism” combines pathways for the biosynthesis of L-methionine, L-cysteine, L-homocysteine, L-homoserine, ethylene, and methanethiol; for degradation of L-serine, L-cysteine, L-methionine, sulfolactate, S-methyl-5’-thioadenosine, and S-methyl-5-thio-alpha-D-ribose 1-phosphate; for homocysteine and cysteine interconversion; and for methionine salvage.
Although KEGG maps (and MetaCyc superpathways) are useful in showing how individual pathways connect and in presenting the larger biochemical context in which a pathway operates, they are not suitable for many types of analyses. For example, using KEGG maps, a program for predicting the metabolic pathways of an organism could not predict methionine biosynthesis independently of ethylene biosynthesis, although many organisms do not produce the latter, because those two separate processes are fused into one KEGG map. Similarly, if a program for enrichment analysis of transcriptomics data detects enrichment of MAP00270, we would not know which actual pathway was in fact enriched: methionine biosynthesis or ethylene biosynthesis. For that matter, MAP00270 could receive a high enrichment score because of differential expression of genes within multiple pathways within that map when in fact no individual pathway was highly enriched. And because maps are so large, a large number of genes must be differentially expressed for a map to obtain a high enrichment score.
The smaller pathways in MetaCyc (and KEGG modules) are advantageous for several reasons: they correspond to a single biological function, the enzymes participating in them are usually regulated as a unit, and they tend to be conserved through evolution. MetaCyc pathway diagrams have several advantages over KEGG modules: MetaCyc diagrams include full chemical compound names and enzyme names (KEGG module diagrams contain only unintelligible identifiers), and MetaCyc diagrams can show the full chemical structures for substrates (the user can select the level of detail shown by choosing to turn on or off the display of structures and/or enzyme names), and can depict regulatory influences on the pathway.
Maps and superpathways are useful in showing how individual pathways connect, and in presenting the larger biochemical context in which a pathway operates. MetaCyc pathways can be displayed at multiple detail levels, such as showing chemical structures for substrates. In addition, all MetaCyc pathway diagrams include chemical names and enzyme names; KEGG module diagrams contain unintelligible identifiers only.
MetaCyc records separately the different pathway variants that have been observed in different organisms. For example, MetaCyc contains six different pathway variants for synthesizing L-lysine. KEGG does not identify pathway variants. Within the large maps defined by KEGG, it is impossible for the user to tell which subnetworks correspond to distinct biological units, nor in which species these units have been elucidated experimentally.
MetaCyc curators author extensive mini-review summaries that describe most pathways and enzymes. KEGG contains short summaries for approximately half of its pathway maps.
MetaCyc pathways are labeled with the name(s) of some of the species in which the presence of those pathways has been experimentally determined, whereas such information is irrelevant for KEGG maps since they are chimeric. Pathways in MetaCyc and in other BioCyc PGDBs contain evidence codes that indicate whether experimental or computational evidence supports the presence of the pathway in that organism; KEGG does not use evidence codes.
MetaCyc proteins contain enzyme properties such as subunit composition, substrate specificity, cofactor requirements, activators, and inhibitors. KEGG has only cofactor data. However, because those data are associated with KEGG reactions rather than with KEGG enzymes, it is difficult to be sure for which proteins from which species the cofactor requirement was experimentally elucidated.
MetaCyc compounds include calculated Gibbs free energies of formation, SMILES, and InChIs, which are missing from KEGG compounds.
Comparing MetaCyc version 27.1 (August 2023) with KEGG version 107.0+ (September 2023), we find that MetaCyc contained contained 3,128 pathways (7.0 times more than KEGG), compared to the 445 metabolic modules in KEGG. MetaCyc contained 18,819 (1.6 times more) reactions, compared to 11,964 in KEGG, and MetaCyc contains 19,173 metabolites compared to 19,136 in KEGG. MetaCyc contains 10,618 textbook-equivalent pages of mini-review summaries for enzymes and pathways versus 1,002 textbook-equivalent pages for KEGG. MetaCyc cites 76,000 publications, from which its curation was drawn (we do not know the equivalent number for KEGG).
BioCyc Organism-Specific PGDBs Compared to KEGG Species Views of Pathway Maps
KEGG version 107.0+ contained 9,313 organisms whereas BioCyc version 27.1 contained 20,042 organism databases (2.2 times more).
Sixty nine of the BioCyc databases are designated as Tier 1 or Tier 2, meaning they have undergone some amount of manual curation. BioCyc curation was drawn from 146,000 publications compared to 71,301 publications cited by KEGG. In some cases, BioCyc databases received several person-years of curation (e.g., for Arabidopsis thaliana) or even person-decades (e.g., for EcoCyc and MetaCyc) of curation. In contrast, KEGG curates only its reference pathway maps and modules; it does not curate organism-specific views of those data, which are generated computationally. Manually curated databases have significant of advantages, including higher accuracy and richer information. Curators remove incorrectly predicted pathways and add pathways that should have been predicted. Curators also add additional information from the literature that often results in modifying the original annotation, and add content such as mini-review summaries, evidence codes, literature citations, and enzyme properties.
Pathway Tools Software Compared to KEGG Software
The Pathway Tools software that underlies MetaCyc and BioCyc is more advanced than the KEGG software in many respects. Pathway Tools can be installed locally at your site, and many of its operations are available via the BioCyc website.
Inference tools: Pathway Tools predicts the following information for an annotated genome: the reactome of the organism, the metabolic pathways of the organism, pathway hole fillers (genes predicted to encode enzymes that catalyze pathway reactions with no attached enzymes), and operons. KEGG offers the first tow functions, but lacks the latter two.
Metabolic flux models: The Pathway Tools MetaFlux component supports creation of quantitative metabolic flux models from Pathway/Genome Databases; KEGG does not offer a similar tool.
Query tools: Pathway Tools has an extensive suite of query tools under its Web Search menu; KEGG lacks most of these tools.
Omics data analysis: Pathway Tools has many omics data analysis tools including:
Painting omics data onto individual pathway diagrams (present in KEGG)
Painting omics data onto multi-pathway diagrams called pathway collages (not present in KEGG)
Painting omics data onto a metabolic network diagram (present in KEGG, but without the animation)
The Omics Dashboard — an innovative tool for analyzing omics data (not present in KEGG)
Comparative analysis tools: Pathway Tools has a number of comparative analysis tools not present in KEGG
Metabolic route search: Pathway Tools can search for routes through a metabolic network connecting a specified starting compound to an ending compound (not present in KEGG).
10.2 EAWAG Biocatalysis/Biodegradation Database
The Biocatalysis/Biodegradation Database was developed by the University of Minnesota and used to be known as UM-BBD. The database contains information on microbial biocatalytic reactions and biodegradation pathways for chemicals largely considered to be potential environmental pollutants.
The data is now hosted at the Swiss Federal Institute of Aquatic Science and Technology and the database is known as the EAWAG Biocatalysis/Biodegradation Database. The web site states that the database has not been updated since 2016. As of January 18, 2023 the database contained 219 pathways and 993 enzymes from 543 microorganisms. Pathways were curated from the biomedical literature and some contain significant comments and literature citations.
10.3 Reactome
Reactome is a curated database of biological processes in humans and a few other organisms. It covers biological pathways ranging from the basic processes of metabolism to high-level processes such as hormonal signaling. Reactome information is curated form the literature, and includes significant comments and literature citations. Reactome contains far fewer metabolic pathways than does MetaCyc, and because most Reactome pathways are curated based on human biology — Reactome does not have the taxonomic breadth of MetaCyc, although Reactome pathways have been computationally projected to a number of model organisms.
11 The MetaCyc Team
This section summarizes the many past and present contributors to the MetaCyc project.
11.1 Current Contributors
Roles: Curation of pathways, software development, Website operations
Peter D. Karp, Ph.D. — Bioinformatics Director
Ron Caspi, Ph.D. — Curator
Anamika Kothari — Programmer
Markus Krummenacker — Bioinformatics Scientist
Suzanne Paley — Computer Scientist
Pallavi Subhraveti — Scientific Programmer
11.2 Past Contributors
Martha Arnaud, Ph.D., formerly at SRI International
Joseph M. Dale, formerly at SRI International
Kate Dreher, Ph.D., International Maize and Wheat Improvement Center, CIMMYT
Hartmut Foerster, Ph.D., Boyce Thomson Institute for Plant Research
Fred Gilham, formerly at SRI International
John Ingraham, Ph.D., UC Davis
A. Karthikeyan, Ph.D, formerly at the Carnegie Institute of Washington, Plant Department
Cindy Krieger, Ph.D., formerly at SRI International
Lukas Mueller, Ph.D., Boyce Thomson Institute for Plant Research
John Pick, formerly at SRI International
Liviu Popescu, formerly at SRI International
Sunita Reddy, formerly at SRI International
Seung Yon (Sue) Rhee, Ph.D., Carnegie Institution for Science, Department of Plant Biology
Monica Riley, Ph.D., Marine Biological Laboratory (deceased)
Alfred Spormann, Ph.D., Stanford University
Christophe Tissier, Ph.D., formerly at the Carnegie Institution for Science, Department of Plant Biology
Alfred Wang, formerly at Stanford University
Deepika Weerasinghe, Ph.D., formerly at SRI
Peifen Zhang, Ph.D., Carnegie Institution for Science, Department of Plant Biology
12 Submitting Pathways for Incorporation into MetaCyc
We would be happy to incorporate pathways created by other scientists into the database.
If you are a Pathway Tools user and have created a pathway that fits our criteria, please send it to us. When we include externally submitted pathways in MetaCyc, we credit the contribution in the MetaCyc release notes, and if you wish, your name and your institution will appear on the pathway page.
In addition, by submitting pathways to MetaCyc you not only add to MetaCyc, but you increase the power of the PathoLogic metabolic-pathway prediction software. PathoLogic recognizes MetaCyc pathways in genome sequence data, and is now in use by hundreds of groups worldwide.
If you would like to submit a pathway for inclusion in a future release of MetaCyc, please make sure that you curate the pathway following these guidelines:
The pathway must be experimentally proven, and must be described in a published journal article
Each pathway and enzyme must have summaries and literature citations
Each pathway gene should have a link to a sequence database (e.g. Entrez); and a citation, if available
Enzymes should have as much information as possible, such as optimal pH and temperature, Km values, inhibitor, activator and cofactor information, etc.
Please include evidence codes for enzymes and pathways
Please make sure to use the Author Crediting feature (found within the Pathway Info Editor) to add the names of the curators and their institution affiliations . This will insure that the appropriate credit for your pathway is published on the BioCyc web page.
For examples of pathways that have been curated based on these guidelines, please see:
Further information can be found in the Curator’s Guide for Pathway/Genome Databases.
12.1 How to Ensure that You and Your Organization Receive the Appropriate Credit
Pathway Tools includes an author crediting system that can attach author and organization credentials to individual pathways. We recommend that prior to creating new objects in the PGDB you should create an Organization frame for your institute and an Author frame for yourself. This way, items that you create afterwards will be associated with these frames, providing you with the credit that you deserve. This credit information would be retained upon exporting the pathways and importing them into MetaCyc. It is also possible to add credit information to older pathways that were created prior to the creation of your author frame, through the Pathway Info Editor.
Detailed instructions on how to create organization and author frames are found in the pathway Tools user manual, in the section “Creating Author Frames”.
12.2 How to Send Pathways to MetaCyc
Pathways should be exported into a text file, which can be emailed to us at: . The procedure for exporting a pathways is:
Display the pathway in Pathway Tools; right click on its name and choose: Edit → Add object to file export list
Repeat until you have added all pathways to be submitted
From the file menu, choose: Export → Selected Objects to Lisp-Format file
Email us the resulting Lisp file
Please indicate if you would like your name and/or affiliation to appear on the pathway and enzyme pages.
13 MetaCyc Publications
If you use MetaCyc in your research, we ask that you cite the following publication:
[MetaCyc20] Caspi, R., Billington, Keseler, I.M., Kothari, A.,
Krummenacker, M., Midford, P.E., Ong, Q., Paley, S., Subhraveti,
P. and Karp, P.D. (2020)
The MetaCyc database of metabolic pathways and enzymes - a 2019 update
Nucleic Acids Research 48(D1):D445–D453.
13.1 Additional Publications
[MetaCyc18] Caspi, R., Billington, Fulcher, C.A., Keseler, I.M., Kothari, A.,
Krummenacker, M., Latendresse, M., Midford, P.E., Ong, Q., Paley, S., Subhraveti, P. and Karp, P.D. (2018)
The MetaCyc Database of metabolic pathways and enzymes
Nucleic Acids Research 46(1):D633-D639.
[MetaCyc16] Caspi, R., Billington, R., Ferrer, L., Foerster, H., Fulcher, C.A., Keseler, I.M., Kothari, A.,
Krummenacker, M., Latendresse, M., Mueller, L.A., Ong, Q., Paley, S., Subhraveti, P., Weaver, D.S. and Karp, P.D.(2016)
The MetaCyc Database of metabolic pathways and enzymes and the BioCyc
collection of Pathway/Genome Databases
Nucleic Acids Research 44(1):D471-D480.
[MetaCyc14] Caspi, R., Altman, T., Billington, R., Dreher, K., Foerster, H, Fulcher, C.A., Holland, T.A., Keseler, I.M., Kothari, A., Kubo, A., Krummenacker, M., Latendresse, M., Mueller, L.A., Ong, Q., Paley, S., Subhraveti, P., Weaver, D.S., Weerasinghe, D., Zhang, P., and Karp, P.D.(2014)
The MetaCyc Database of metabolic pathways and enzymes and the BioCyc
collection of Pathway/Genome Databases
Nucleic Acids Research 42(1):D459-D471.
[MetaCyc13] Altman, T., Travers, M., Kothari, A., Caspi, R. and Karp, P.D.
A systematic comparison of the MetaCyc and KEGG pathway
databases
BMC Bioinformatics 14:112 2013
[Curation13] Caspi, R., Dreher, K, and Karp, P.D.
The challenge of constructing, classifying and representing metabolic pathways
FEMS Microbiology Letters 345:85 2013
[MetaCyc12] Caspi, R., Altman, T., Dreher, K., Fulcher, C.A., Subhraveti, P., Keseler, I.M., Kothari, A., Krummenacker, M., Latendresse, M.,
Mueller, L.A., Ong, Q., Paley, S., Pujar, A., Shearer, A.G., Travers, M., Weerasinghe, D., Zhang, P., and Karp, P.D. (2012)
The MetaCyc Database of metabolic pathways and enzymes and the BioCyc
collection of Pathway/Genome Databases
Nucleic Acids Research 40(1):D742-D753 2012.
[MetaCyc11] Karp, P.D., and Caspi, R.,
A survey of metabolic databases emphasizing the MetaCyc family
Archives of Toxicology 85:1015–33 2011.
[MetaCyc10] Caspi, R., Altman, T., Dale, J.M., Dreher, K., Fulcher, C.A., Gilham, F., Kaipa, P., Karthikeyan, A.S., Kothari, A., Krummenacker, M., Latendresse, M., Mueller, L.A., Paley, S., Popescu, L., Pujar, A., Shearer, A., Zhang, P. and Karp, P.D. (2010)
The MetaCyc Database of metabolic pathways and enzymes and the BioCyc
collection of Pathway/Genome Databases
Nucleic Acids Research 38(1):D473-D479.
[MetaCyc08] Caspi, R., Foerster, H., Fulcher, C.A., Kaipa, P., Krummenacker, M.,
Latendresse, M., Paley, S., Rhee, S.Y., Shearer, A., Tissier, C.,
Walk, T.C., Zhang, P. and Karp, P.D. (2008)
The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases
Nucleic Acids Research 36(1):D623–D631.
[MetaCyc06] Caspi, R., Foerster, H., Fulcher, C.A., Hopkinson, R.,
Ingraham, J., Kaipa, P., Krummenacker, M., Paley, S., Pick, J.,
Rhee, S.Y., Tissier, C., Zhang, P. and Karp, P.D. (2006)
MetaCyc:
A multiorganism database of metabolic pathways and enzymes
Nucleic Acids Research, 34:D511–D516.
[MetaCyc04]
Krieger, C.J., Zhang, P., Mueller, L.A., Wang, A., Paley, S.,
Arnaud, M., Pick, J., Rhee, S.Y., and Karp, P.D. (2004)
MetaCyc: A Multiorganism Database of Metabolic Pathways and Enzymes,
Nucleic Acids Research 32(1):D438–42.
[MetaCyc03]
Karp, P.D. (2003)
The MetaCyc Metabolic Pathway Database
In: Metabolic Engineering, B. Kholodenko and H. Westerhoff eds., Horizon Scientific Press.
[MetaCyc02]
Karp, P.D., Riley, M., Paley, S. and Pellegrini-Toole, A. (2002)
The MetaCyc Database
Nucleic Acids Research, 30(1):59–61.
[MetaCyc00] Karp, P.D., Riley, M., Saier, M., Paulsen, I.T., Paley, S., and Pellegrini-Toole, A. (2000)
The EcoCyc and MetaCyc Databases
Nucleic Acids Research, 28(1):56–59.
See also the BioCyc Publications Page.
14 How to Learn More
References
| [1] |
R. Caspi, R. Billington, C. A. Fulcher, I. M. Keseler, A. Kothari,
M. Krummenacker, M. Latendresse, P. E. Midford, Q. Ong, W. K. Ong, S. Paley,
P. Subhraveti, and P. D. Karp.
The MetaCyc database of metabolic pathways and enzymes.
Nuc Acids Res, 46(D1):D633–9, 2018.
https://doi.org/10.1093/nar/gkx935,.
|
| [2] |
R. Caspi, R. Billington, L. Ferrer, H. Foerster, C. A. Fulcher, I. M. Keseler,
A. Kothari, M. Krummenacker, M. Latendresse, L. A. Mueller, Q. Ong, S. Paley,
P. Subhraveti, D. S. Weaver, and P. D. Karp.
The MetaCyc database of metabolic pathways and enzymes and the
BioCyc collection of Pathway/Genome Databases.
Nuc Acids Res, 44(D1):D471–80, 2016.
https://doi.org/10.1093/nar/gkv1164.
|
| [3] |
R. Caspi, T. Altman, R. Billington, K. Dreher, H. Foerster, C.A. Fulcher, T.A.
Holland, I.M. Keseler, A. Kothari, A. Kubo, M. Krummenacker, M. Latendresse,
L.A. Mueller, Q. Ong, S. Paley, P. Subhraveti, D.S. Weaver, D. Weerasinghe,
P. Zhang, and P. D. Karp.
The MetaCyc database of metabolic pathways and enzymes and the
BioCyc collection of Pathway/Genome Databases.
Nuc Acids Res, 42:D459–71, 2014.
https://doi.org/10.1093/nar/gkt1103.
|
| [4] |
R. Caspi, T. Altman, K. Dreher, C.A. Fulcher, P. Subhraveti, I. Keseler,
A. Kothari, M. Krummenacker, M. Latendresse, L.A. Mueller, Q. Ong, S. Paley,
A. Pujar, A.G. Shearer, M. Travers, D. Weerasinghe, P. Zhang, and P. D. Karp.
The MetaCyc database of metabolic pathways and enzymes and the
BioCyc collection of Pathway/Genome Databases.
Nuc Acids Res, 40:D742–53, 2012.
https://doi.org/10.1093/nar/gkr1014.
|
| [5] |
P. D. Karp and R. Caspi.
A survey of metabolic databases emphasizing the MetaCyc family.
Arch of Toxicol., 85:1015–33, 2011.
http://www.springerlink.com/content/1u7l1127766194w3/.
|
| [6] |
R. Caspi, T. Altman, J.M. Dale, K. Dreher, C.A. Fulcher, F. Gilham, P. Kaipa,
A.S. Karthikeyan, A. Kothari, M. Krummenacker, M. Latendresse, L.A. Mueller,
S. Paley, L. Popescu, A. Pujar, A.G. Shearer, P. Zhang, and P. D. Karp.
The MetaCyc database of metabolic pathways and enzymes and the
BioCyc collection of Pathway/Genome Databases.
Nuc Acids Res, 38:D473–9, 2010.
https://doi.org/10.1093/nar/gkp875.
|
| [7] |
R. Caspi, H. Foerster, C.A. Fulcher, P. Kaipa, M. Krummenacker, M. Latendresse,
S. Paley, S. Y. Rhee, A. Shearer, C. Tissier, T.C. Walk, P. Zhang, and P. D.
Karp.
The MetaCyc database of metabolic pathways and enzymes and the
BioCyc collection of Pathway/Genome Databases.
Nuc Acids Res, 36:D623–31, 2008.
https://doi.org/10.1093/nar/gkm900.
|
| [8] |
R. Caspi, H. Foerster, C.A. Fulcher, R. Hopkinson, J. Ingraham, P. Kaipa,
M. Krummenacker, S. Paley, J. Pick, S. Y. Rhee, C. Tissier, P. Zhang, and
P. D. Karp.
MetaCyc: A multiorganism database of metabolic pathways and
enzymes.
Nuc Acids Res, 34:D511–6, 2006.
https://doi.org/10.1093/nar/gkj128.
|
| [9] |
C.J. Krieger, P. Zhang, L. A. Mueller, A. Wang, S. Paley, M. Arnaud, J. Pick,
S. Y. Rhee, and P. D. Karp.
MetaCyc: A multiorganism database of metabolic pathways and
enzymes.
Nuc Acids Res, 32:D438–42, 2004.
https://doi.org/10.1093/nar/gkh100.
|
| [10] |
P. D. Karp.
The MetaCyc metabolic pathway database.
In Metabolic Engineering. Horizon Scientific Press, 2003.
|
| [11] |
P. D. Karp, M. Riley, S. Paley, and A. Pellegrini-Toole.
The MetaCyc database.
Nuc Acids Res, 30(1):59–61, 2002.
https://doi.org/10.1093/nar/30.1.59.
|
| [12] |
E. W. Sayers, T. Barrett, D. A. Benson, S. H. Bryant, K. Canese, V. Chetvernin,
D. M. Church, M. DiCuccio, R. Edgar, S. Federhen, M. Feolo, L. Y. Geer,
W. Helmberg, Y. Kapustin, D. Landsman, D. J. Lipman, T. L. Madden, D. R.
Maglott, V. Miller, I. Mizrachi, J. Ostell, K. D. Pruitt, G. D. Schuler,
E. Sequeira, S. T. Sherry, M. Shumway, K. Sirotkin, A. Souvorov,
G. Starchenko, T. A. Tatusova, L. Wagner, E. Yaschenko, and J. Ye.
Database resources of the National Center for Biotechnology
Information.
Nuc Acids Res, 37(Database issue):D5–15, 2009.
|
| [13] |
S. E. Stein, S. R. Heller, and D. Tchekhovskoi.
An open standard for chemical structure representation: The IUPAC
chemical identifier.
In Proc. 2003 International Chemical Information Conference
(Nimes), pages 131–43, 2003.
|
| [14] |
M. Latendresse, J.P. Malerich, M. Travers, and P. D. Karp.
Accurate atom-mapping computation for biochemical reactions.
J Chem Inf Model, 52(11):2970–82, 2012.
|
| [15] |
T. Altman, M. Travers, A. Kothari, R. Caspi, and P. D. Karp.
A systematic comparison of the MetaCyc and KEGG pathway
databases.
BMC Bioinformatics, 14:112, 2013.
http://www.biomedcentral.com/1471-2105/14/112/abstract.
|